Data
This view provides an interface to the HitWalker program which ranks genes containing variants with respect to functional data or supplied gene sets.
The random walker restart probability is a value between 0 – 1 which controls the 'importance' of the functional hits/supplied genes. Higher values will progressively increase the score for 'closer' genes in a network context at the expense of those further away.
Gene-gene interaction data is currently derived from Reactome Functional Interactions. See Wu G, Feng X, Stein L.
Genes mutations can be prioritized based on several choices for scoring genes with respect to inhibitor screens.
Subnetworks are generated by finding and aggregating a series of unweighted steiner trees derived from the union of the desired number of top gene score and mutation genes. Based on a re-implementation of the STM method of Sadeghi and Frohlich .
Setting a seed allows for reproducing network figures derived from STM.
Visuals
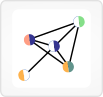
Subnetworks are shown with genes as text labels and interactions as lines between them.
If a sample has inhibitor gene score data for a gene, that data is shown in color-coded semicircles underneath the gene label.
Mutations in a gene for the currently selected sample are shown with a purple circle at the end of the gene label, while mutations from any sample are shown with orange circles.
User interactions
Within the subnetwork plot, variants in a gene can be shown/hidden with the "Show variants" checkbox.
Genes can be dragged to reposition and pin them in place.