Variant Filters
Vizome allows you to work with the whole set of somatic variants in our BeatAML dataset by default.
But you may want to apply a filter so that you're working with only a subset of the variants.
Vizome presents six ways to filter variants. You may combine several filters, if desired.
Variant-filtering options include:
- Custom variant sets
- Cohort variant frequency
- Number of reads
- Variant allele frequency
- Variant type
- Presence/absence in samples/databases
You may also expand the set of variants you're working with by setting the "Show variants excluded from analysis" filter. This will add back in variants that were excluded from analysis.
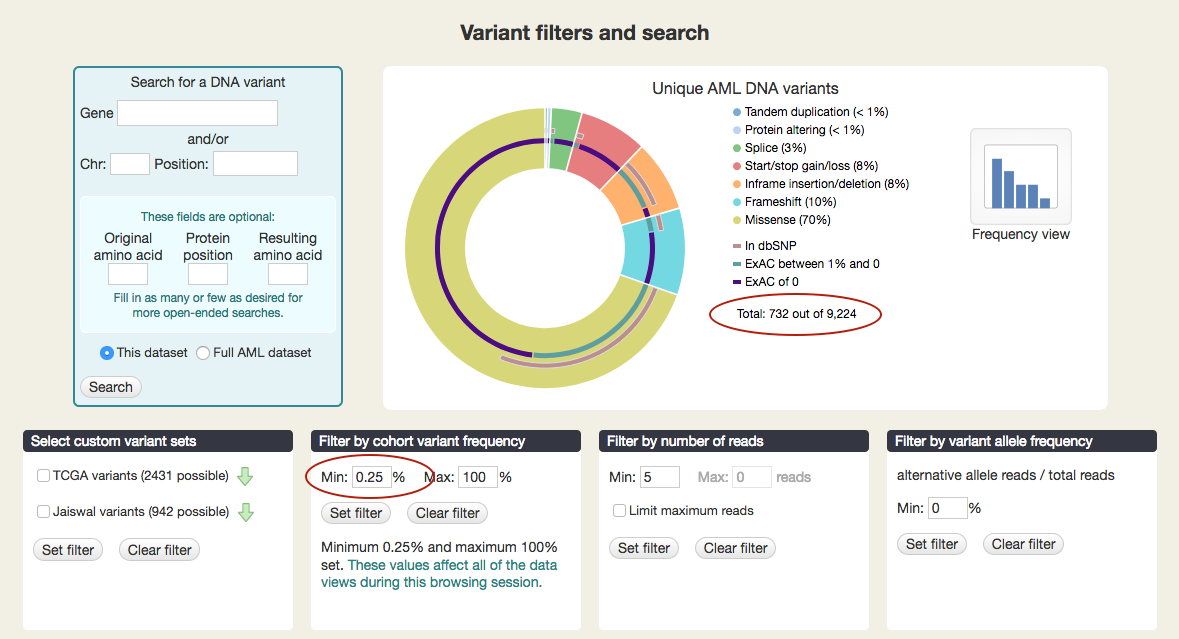
As you set these filters, you will see the number of unique variants change in the figure at the top. When no filters are set, you'll see: "Total: 4,839 out of 9,224." (The 9,224 total includes variants excluded from analysis.) Note: you will not see the total number change when you set a minimum read or a variant allele frequency filter. Unique variants do not have those values, as they are not linked to individual samples.
For example, setting the minimum cohort variant frequency to 0.25 leaves only 732 variants, out of 9,224.
You will also see a notification appear at the top of the page, summarizing the current set of global filters.
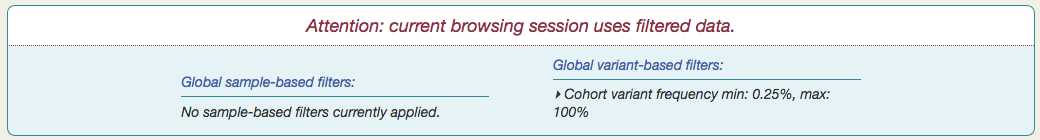
Any filters you set here will affect what variants you see throughout Vizome, for the current browsing session.