Data
This view provides an interface to the HitWalker program which ranks genes containing variants with respect to functional data or supplied gene sets.
The random walker restart probability is a value between 0 – 1 which controls the 'importance' of the functional hits/supplied genes. Higher values will progressively increase the score for 'closer' genes in a network context at the expense of those further away.
There are two types of gene-gene interaction data that can current be used:
1) A set of directed interactions derived from multiple pathway databases found in Pathway Commons, a gene will rank highly here if it is 'upstream' of one or more of the functional hits or supplied genes.
2) The 'classical' set of undirected STRING interactions, a gene ranks highly here if it is close to the hits / specified genes without respect to directionality.
Genes can be chosen based on whether they are siRNA and/or gene score hits as well as user supplied gene symbols. Symbols and siRNA hits are treated as having the highest observed gene score value for a given sample.
Subnetworks are generated by finding and aggregating a series of unweighted steiner trees encompassing the desired number of hit genes and variant genes. This is done separately for both the STRING and Pathway Commons interaction data and the merged result is provided to the user.
Visuals
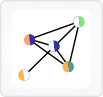
Subnetworks are shown with genes as text labels and interactions as lines between them. The lines can vary in thickness, with a thicker line indicating that an interaction was found in both STRING and Pathway Commons, and can also be solid or dotted, indicating that the interaction was found in Pathway Commons or STRING only, respectively.
If a sample has siRNA z-score and/or inhibitor gene score data for a gene, that data is shown in color-coded semicircles underneath the gene label.
Variants in a gene for the currently selected sample are shown with a purple circle at the end of the gene label, while variants from any sample are shown with orange circles.
User interactions
Within the subnetwork plot, variants in a gene can be shown/hidden with the "Show variants" checkbox.
Genes can be dragged to reposition and pin them in place.
To release a pinned node, click its enclosing square.